Exploring Experimental Hematology: May 2020 (Volume 85)
Exploring Experimental Hematology: Single-cell transcriptome in chronic myeloid leukemia: pseudotime analysis reveals evidence of embryonic and transitional stem cell states
In this issue of Simply Blood, David Stachura is exploring Experimental Hematology by highlighting and deconstructing one of the journal’s latest manuscripts by first author Sarah Pagliaro, from the laboratory of Ali G. Turhan [https://www.exphem.org/article/S0301-472X(20)30142-9/].
Reasons you should read this paper:
About 15% of adult leukemia is categorized as Chronic Myeloid Leukemia (CML). It’s a clonal hematopoietic disease that usually results from a t(9;22) chromosomal translocation, causing the formation of the BCR-ABL oncogene. This tyrosine kinase (TK) is responsible for activating downstream molecular pathways that allow the uncontrollable growth of cancer cells. While tyrosine kinase inhibitor (TKI) drugs that can slow and stop the progression of the disease exist, many patients become immune to these therapies as the clonal expansion of drug-resistant leukemic stem cells (LSCs) occurs.
In this manuscript, Pagliaro et al., asked the question: “If at the time of diagnosis of CML there is a population of heterogeneous cancer cells, could one examine these individual cells and see what genes are present that may contribute to oncogenesis?”. What they found was that a panel of genes important for pluripotency (including NANOG, OCT4, and SOX2) were expressed. They also found that CML cells are not static; they have a dynamic gene expression pattern. Importantly, many of these molecular pathways can be targeted with directed therapeutics. This indicates that combining TKIs with other drugs that block these pathways may be an effective treatment for CML.
Strategy used in this paper:
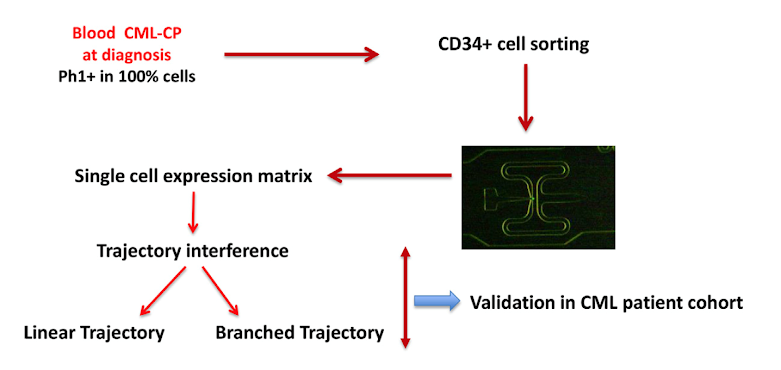
Figure 1: General schematic for single cell analysis
In general, the authors utilized Fluidigm technology to analyze the expression of genes in single CD34+ cells isolated from CML patients. They also utilized a bioinformatics tool called “pseudotime”, which is a way to quantitatively measure biological progression through processes like hematopoietic cell differentiation.
Figure 2: Pluripotency genes are highly expressed in cells from patients (UPN1, 2, and 3)
My reason for reading this paper:
When imatinib came on the market in the early 2000’s, it was a transformational drug. This specific small molecule targeting the BCR-ABL oncogene has saved many thousands of lives over the past 15+ years. However, patients can still relapse and become immune to the drug (and many of the second- and third-generation TKIs). So, knowing all the molecular pathways to target in CML is essential. And, if there are a mix of CML cells that are potentially activated in different ways, different drugs are likely needed to stop oncogenesis. This manuscript gets to the heart of that issue, answering some important questions about what molecular pathways we should be targeting combinatorially to save more lives in the future. As my laboratory also studies CML, I am always interested in new ideas and thoughts to target this disease.
Written by:
David Stachura, PhD
ISEH Publications Committee Member
Assistant Professor
Department of Biological Sciences
California State University, Chico
dstachura@csuchico.edu
myweb.csuchico.edu/~dstachura/Stachura_Laboratory
https://www.facebook.com/StachuraZebrafishLab/
ISEH Publications Committee Member
Assistant Professor
Department of Biological Sciences
California State University, Chico
dstachura@csuchico.edu
myweb.csuchico.edu/~dstachura/Stachura_Laboratory
https://www.facebook.com/StachuraZebrafishLab/



.png)


Comments
Post a Comment